
Welcome to the Population Genomics Laboratory of Department of Genetics, Genomics and Informatics at the University of Tennessee Health Science Center. We are interested in understanding causes and consequences of genetic diversity and how natural selection in humans affects loci related to diseases.
Fascinating (Lieutenant Spock)

Research
Admixture Mapping and Pangenomics Analysis in the BIG Initiative Cohort
Improving Phenotype-Genotype Linkages through Ancestry Patterns
The Biorepository and Integrative Genomics (BIG) Initiative in Tennessee has developed a pioneering resource to address gaps in genomic research by linking genomic, phenotypic, and environmental data from a diverse Mid-South population, including underrepresented groups.
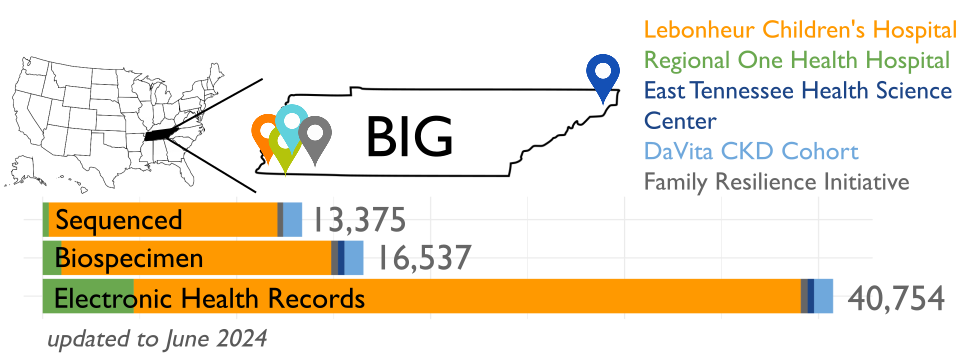
BIG is recruiting participants from Memphis, TN, with plans to include a total of 100,000 samples over the next five years and it is partnering with the Genomic Information Commons, a consortium of top children’s hospitals, to conduct genomics research aimed at discovering the genetic foundations of human disease in diverse populations.
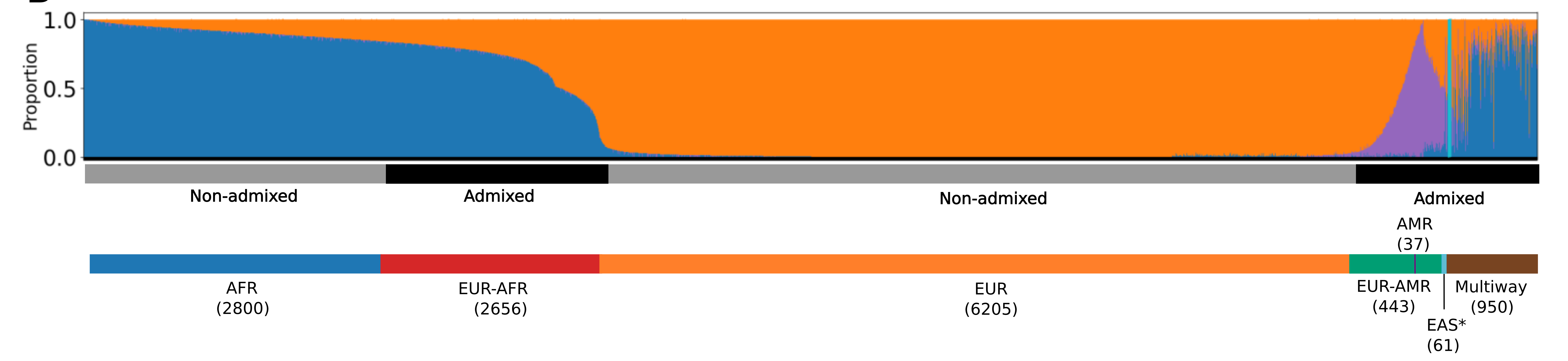
Analysis of 13,152 genomes from BIG revealed significant genetic diversity, with 50% of participants inferred to have non-European or various types of admixed ancestry, highlighting the importance of inclusive genomics in understanding health disparities.
Human Pangenomics
A pangenome is a comprehensive collection of all the genetic variation present in a species, which overcomes the limitations of reference-based genomics by including both common and rare genetic variations in a single reference genome.
The Human Pangenome Reference Project aims to sequence 300 people to create a pangenome of 600 haplotypes and has currently released a first draft of the human pangenome reference based on 47 phased diploid assemblies from a group of genetically diverse individuals
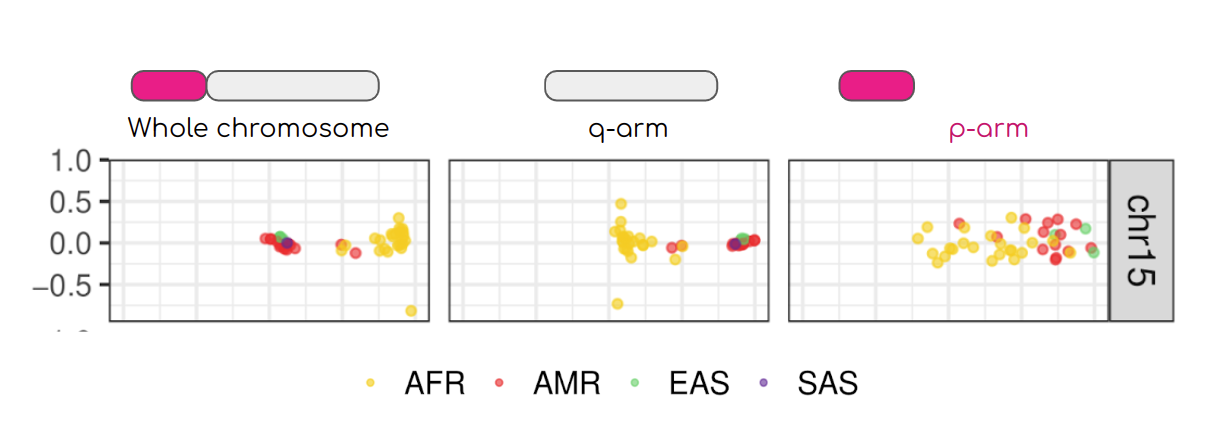
My team contributed to evaluate, for the first time, human population structure using markers from short arms of the acrocentric chromosomes [PMID: 37165242]. We found that markers from these regions have less power to distinguish populations compared to other regions. This is consistent with the understanding that short arms of acrocentric chromosomes undergo recombination between non-homologous chromosomes, similar to the X and Y pseudohomologous regions [PMID: 37165241]. Our findings on the patterns of linkage disequilibrium in these regions support this idea.
Funding
-
NIH U01HG013760 - Building Tools and Community to Make Pangenomes Accessible
Mouse Pangenomics
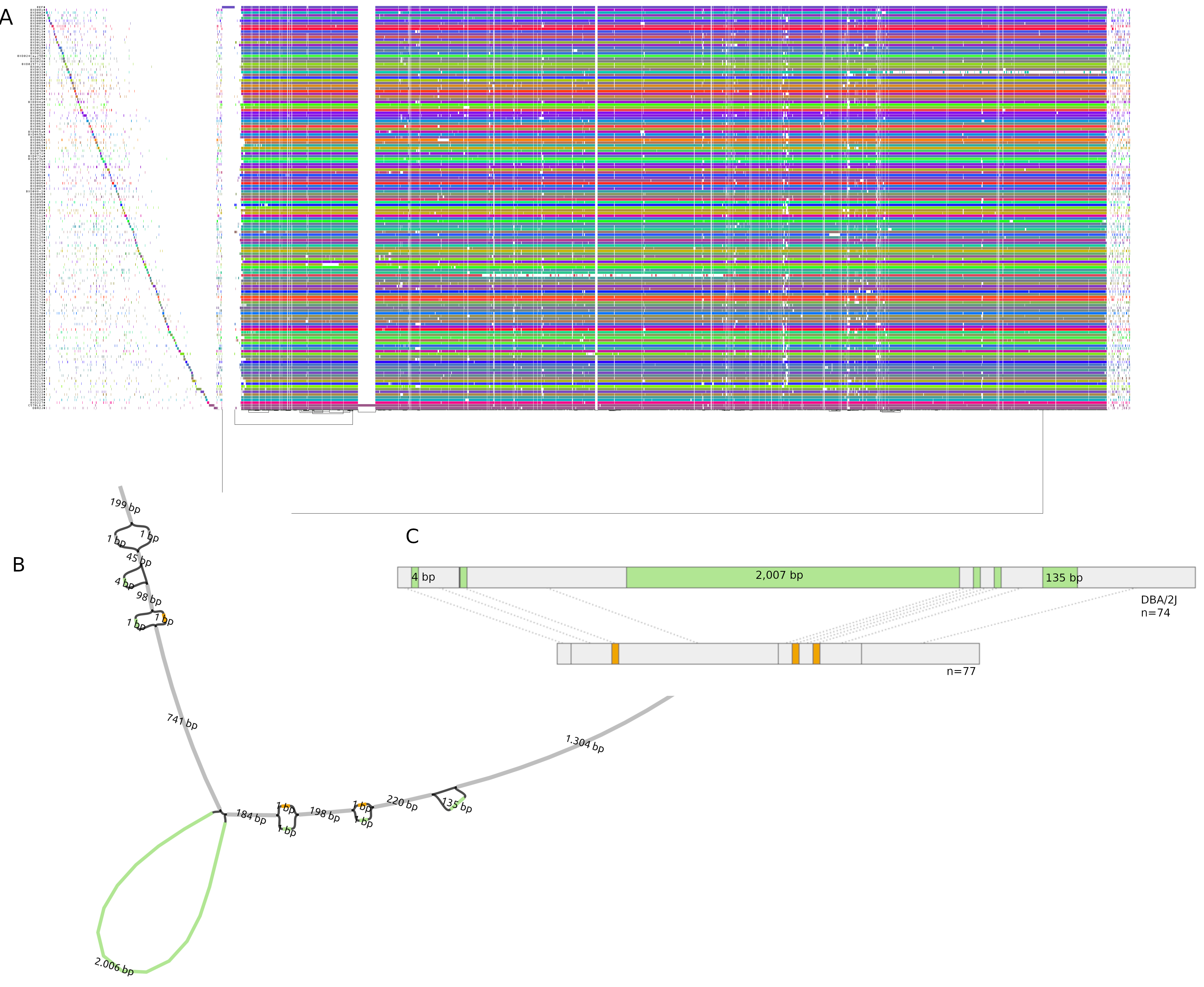
Mice members of BXDs family have been inbred for 20-200 generations. They are of great value for mapping complex traits and phenome-wide association analyses. Current genomic studies on BXD assume a single linear reference genome, making it difficult to observe sequences diverging from the reference, therefore limiting the accuracy and completeness of analyses. Pangenome models overcome this limitation as they contain the full genomic information of a species.
We are building a reference pangenome for all extant members of all BXD families leveraging third generation and 10X sequence data. We will analyze the genetic variation in relation to thousands of phenotypes in the genenetwork.org/ database.
Genomics of early embryonic development
How natural selection acts on early human development
We investigate adverse outcomes of embryonic development like recurrent pregnancy loss and preeclampsia to identify the genetic factors that influence reproductive outcomes and pregnancy complications. This knowledge furthers our understanding of human evolution and informs efforts to improve pregnancy outcomes.
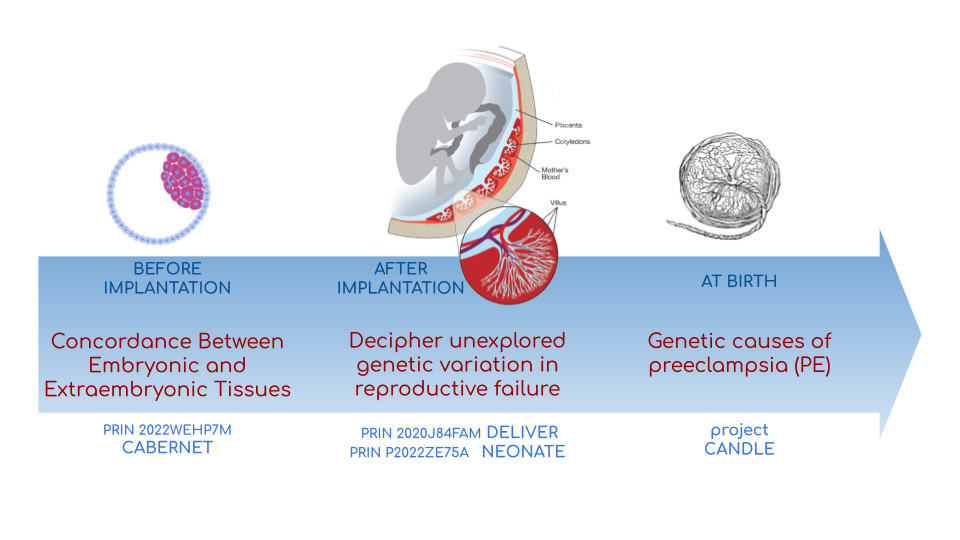
In CABERNET we aim to determine the extent of chromosomal mosaicism between embryonic and extraembryonic tissues using single cell DNA sequencing.
In DELIVER / NEONATE we want to identify genetic factors contributing to reproductive failure and recurrent miscarriage. We will use single cell strand sequencing to map balanced rearrangements and whole genome sequencing of euploid miscarried embryos to identify causative variants.
In CANDLE we want to uncover gene expression patterns associated with APOL1 risk alleles and preeclampsia in African American women. We will examine the role of ancestry in mediating the relationship between APOL1 genotype and preeclampsia risk. The results can provide insights into genetic and molecular basis of preeclampsia.
-
Francesca Antonacci University of Bari Aldo Moro
-
Carlo Alviggi, University of Naples Federico II
-
Antonio Lamarca, University of Modena and Reggio Emilia
-
Marcella Vacca, National Research Council
-
Gabriella Lania, National Research Council
Funding
-
PRIN 2020J84FAM Ministero dell’Universita e della Ricerca
-
PRIN 2022WEHP7M Ministero dell’Universita e della Ricerca
-
PRIN P2022ZE75A Ministero dell’Universita e della Ricerca
-
POR Campania FSE 2014-2020 ASSE III – Ob. Sp. 14


People
Vincenza Colonna
Associate Professor, Department of Genetics, Genomics and Informatics website
University of Tennessee Health Science Center, Memphis, TN
Director, Integrative Genomics Biorpository, Department of Pediatrics
Children’s Foundation Research Institute, Memphis, TN
Researcher (on leave of absence), National Research Council website
Institute of Genetics and Biophysics, Naples, Italy

I am a genomicist and an expert in human evolutionary and population genomics and bioinformatics. In my postdoctoral research I was part of the international consortium 1000 Genomes[PMID: 26432245; 23128226] where I led contributions to two specific aspects. First, I contributed to develop FunSeq [PMID: 24092746], a tool that integrates non-coding information from relevant biological databases for the functional characterization of non-coding variants. Second, I lead a genome-wide scan to identify genomic regions with exceptionally high levels of population differentiation [PMID: 24980144] demonstrating that these regions are enriched for positive selection events and that one half may be the result of classic selective sweeps. Findings from both sub-projects have since been applied to demographic inference and the molecular diagnosis of cancer and myeloid malignancies [PMID: 27121471, 22446628], and to deeper studies on positive selection at the ABCA12 gene [PMID: 30890716].
During my PhD I worked on human isolated populations contributing to characterize several isolated populations, describing the genomic consequences of isolation [PMID: 17476112, 19550436, 22713810], contributing to genetic association studies [PMID: 16611673, 18162505] and to characterize rare variation [PMID: 28643794]
I founded and led OBiLab, a project on training in Bioinformatics
Silvia Buonaiuto, Postdoctoral fellow

I work on the admixture mapping and pangenomics analysis in the BIG project. My role involves establishing connections between genotypes and phenotypes especially in admixed individuals through admixture mapping analysis. Additionally, I am leading sequence analysis for the projects related to early embryonic development.
Franco Marsico, Postdoctoral fellow

I am currently working on Admixture Mapping and Pangenomics Analysis in the Biorepository and Integrative Genomics (BIG) Initiative Cohort project. My focus is on studying recent natural selection signals in admixed populations. Additionally, I have a deep interest in evolution and how to compute processes that shape the history of life.
Flavia Villani, PhD Student
I am working on the Mouse Pangenomics Project. My research aims to construct the pangenome graph of model organisms, specifically inbred mice and rats, using a combination of short and long-read sequence data. This will enable genome-wide association studies to be performed directly on the pangenome. Additionally, I have a strong interest in understanding how mobile genetic elements have driven genome evolution through various mechanisms.
Ernestine Amos-Abanyie, PhD Student

I am studying mitochondrial DNA Variation in disease susceptibility in the BXD Mice and the BIG cohort.
Donna Vo, Research associate

I am in charge of sample collection and processing for the Integrative Genomics Biorepository that supports the BIG project.
Visiting
Laura Pignata, Postdoctoral fellow

I am working on the analysis of long-read sequencing data on human samples with mosaic methylation defects. The aim of this study is to identify the mechanisms underlying the methylation defects by examining the role of de novo mutations in cis and somatic recombination events, possibly arisen during early embryogenesis.
Dario Cannella, PhD Student

I am working on a project that leverages pangenomics to uncover the mechanisms behind the evolution and selection of structural genomic diversity across different loci. Additionally, my PhD project is focuses on understanding the relationship between Down Syndrome and its comorbidities, with the goal of enabling a deeper stratification of individuals affected by this common condition.
Angela Carfora, PhD Student

My doctoral research focuses on the genetic adaptations of plants in urban environments, with Raphanus raphanistrum (wild radish) as a model species. Initially, I investigated differential gene expression between urban and extra-urban populations. Currently, my work involves the study of genomic variations through developing a pangenome for R. raphanistrum. This research is being conducted in collaboration with Dr. Colonna’s laboratory at the University of Tennessee Health Science Center in Memphis, where I will undertake part of my studies.
Former members
-
Gianluca Damaggio, Master student, PhD student, 2019-2023
-
Rosanna Maione, Research associate 2023
-
Madeleine Emms, Postdoctoral fellow, 2022-2023
-
Marialaura Zitiello, Master Student, 2022-2023
-
Antonella Mecca, Master Student, 2022-2023
-
Angela Sequino, Master Student, 2022-2023
-
Davide D’angelo, Visiting master student, 2022
-
Giuliana D’Angelo, Master Student, 2019-2020
-
Roberto Sirica, PhD student, 2015-2018
-
Gaia Leandra Cecere, undergraduate student, 2018
-
Marianna Buonaiuto, visiting Postdoc, 2017
-
Lucia De Martino, visiting master Student, 2016
Publications
See them on Google Scholar
Past Research
At this link is possible to found our past Research
Contacts
Vincenza Colonna, PhD
-
University of Tennessee Health Science Center, TSRB room 405
71 S Manassas St, Memphis TN 38163 map -
Children’s Foundation Research Institute
50 N. Dunlap St. Memphis, TN 38105 map -
Istituto di Genetica e Biofisica "Adriano Buzzati-Traverso" piano R, stanza 11
via Pietro Castellino 111 - 80131 Napoli - Italy map